libct
A fast approximate primal-dual solver for tracking-by-assignment cell-tracking problems
Synopsis
![]()
Cell tracking plays a key role in discovery in many life sciences, especially in cell and developmental biology. So far, in the most general setting this problem was addressed by off-the-shelf solvers like Gurobi, whose run time and memory requirements rapidly grow with the size of the input. In contrast, for this solver the growth is nearly linear. Compared to solving the problem with Gurobi, we observe an up to 60 times speed-up, while reducing the memory footprint significantly.
For more a detailed report please see our AISTATS 2020 publication.
Publication and Citation
The creation of this solver was joint effort of multiple persons and research institutes. To cite our this work please use:
S. Haller, M. Prakash, L. Hutschenreiter, T. Pietzsch, C. Rother, F. Jug, P. Swoboda, B. Savchynskyy. A Primal-Dual Solver for Large-Scale Tracking-by-Assignment. AISTATS 2020. [pdf]
@InProceedings{haller20,
title = {A Primal-Dual Solver for Large-Scale Tracking-by-Assignment},
author = {Haller, Stefan and Prakash, Mangal and Hutschenreiter, Lisa and Pietzsch, Tobias and Rother, Carsten and Jug, Florian and Swoboda, Paul and Savchynskyy, Bogdan},
booktitle = {Proceedings of the Twenty Third International Conference on Artificial Intelligence and Statistics},
pages = {2539--2549},
year = {2020},
editor = {Chiappa, Silvia and Calandra, Roberto},
volume = {108},
series = {Proceedings of Machine Learning Research},
address = {Online},
month = {26--28 Aug},
publisher = {PMLR},
pdf = {http://proceedings.mlr.press/v108/haller20a/haller20a.pdf},
url = {http://proceedings.mlr.press/v108/haller20a.html},
}
libct – The Solver Library
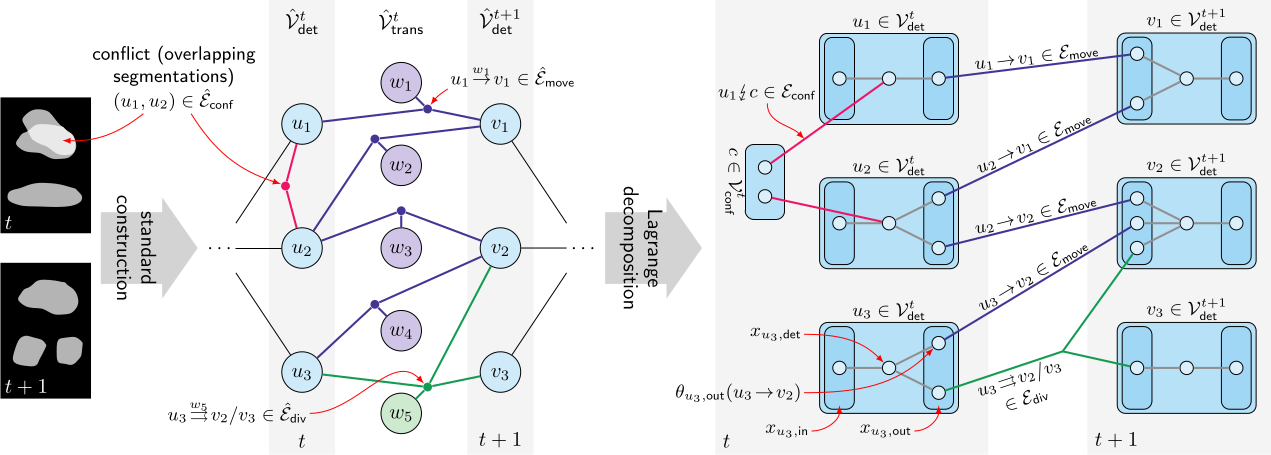
We implemented the suggested solving scheme in a modern C++ library. The source code of this implementation is publicly available and we plan to incorporate further improvements in the future. To make the results presented in this paper reproducible, the repository holding the source code also contains a fixed version which we used during the preparation of the aforementioned publication.
Along with the library we provide Python 3 bindings which allow to feed a text file representation of cell-tracking problems into the native library to run the solver.
Additional information to various aspects of our implementation are available here:
- Source Code Repository
- Invocation and Usage Instructions
- Specification of Input File Format
- Model Generation from Raw Data
Datasets
The generated model files that have been used for preparation of our AISTATS2020 2020 submissions are available here: AISTATS 2020 Models